Live Cell Imaging on Single Cell Arrays
(LISCA)
(LISCA)
Judith A. Müller, Gerlinde Schwake & Joachim O. Rädler
Einzelzellmikroskopie im Hochdurchsatz auf Mikrostrukturen
Biospektrum 28 (2022)
Cellular response to perturbations, if examined at the single cell level, exhibits cell-to-cell differences, distinct dynamic behavior and correlations meaningful for the study of regulatory networks and information processing. Live-cell imaging on single cell arrays (LISCA) facilitates automated acquisition of individual time courses with sharp temporal resolution. Here we discuss expression dynamics after transient GFP transfection and event-time correlations in nanoparticle induced apotosis.
A. Reiser, D. Woschée, S. M. Kempe, J. O. Rädler
JoVe Journal (2021)
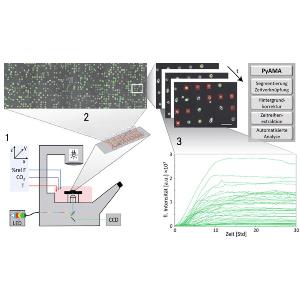
Live-cell Imaging of Single-Cell Arrays (LISCA) is a versatile method to collect time courses of fluorescence signals from individual cells in high throughput. In general, the acquisition of single-cell time courses from cultured cells is hampered by cell motility and diversity of cell shapes. Adhesive micro-arrays standardize single-cell conditions and facilitate image analysis. LISCA combines single-cell microarrays with scanning time-lapse microscopy and automated image processing. Here, we describe the experimental steps of taking single-cell fluorescence time courses in a LISCA format. We transfect cells adherent to a micropatterned array using mRNA encoding for enhanced green fluorescent protein (eGFP) and monitor the eGFP expression kinetics of hundreds of cells in parallel via scanning time-lapse microscopy. The image data stacks are automatically processed by newly developed software that integrates fluorescence intensity over selected cell contours to generate single-cell fluorescence time courses. We demonstrate that eGFP expression time courses after mRNA transfection are well described by a simple kinetic translation model that reveals expression and degradation rates of mRNA. Further applications of LISCA for event time correlations of multiple markers in the context of signaling apoptosis are discussed.
PyAMA is a Python-based Automated Microstructure Analysis tool designed at our chair that can read and display single-cell fluorescence trajectories from time-lapse movie files. Once the background correction and cell tracking have been performed, cells and their corresponding fluorescence traces can be individually monitored and selected or de-selected from the final data evaluation. The most recent and publicly available source code can be downloaded from Github.
Fluorescence fitting
With the Python-based fitting algorithm the resulting single fluorescence traces from PyAMA can be fitted to a translation model based on chemical rate equations as described by Reiser et al. This model uses a three stages approach to describe translation and maturation.
S. Westermayer, J. Mergerle, G. Fritz, U. Gerland & J. O. Rädler
Bacteria can rapidly react to environmental changes by adapting gene expression of certain genes. We investigate the time-dependent response of sugar utilization systems in Escherichia coli on the single-cell level. We study both individual systems and the situation of two competing sugar utilization systems, with the goal to characterize properties that may have been optimized by evolution. Using microfluidic set-ups we expose bacterial cultures to systematically variable environments and use time-lapse microscopy and single cell tracking to acquire single-cell expression kinetics. In mathematical models using cost-benefit analysis and game theoretical concepts, we compare different regulatory schemes as strategies to cope with variable environments.
This work is supported by the DFG through the priority program SPP